How to Use CHITRA
Step-by-step guide to using CHITRA and exploring example datasets
HOW TO USE CHITRA?
Below are the step-by-step instructions, accompanied by pictorial representations, to ensure a seamless experience with CHITRA.
Step 1: Getting Started
- On the landing page, click the “Get Started” button to begin.
Step 2: Uploading Files or Viewing Example Data
- On the next page, you can either:
- Upload Files
- View Example Datasets
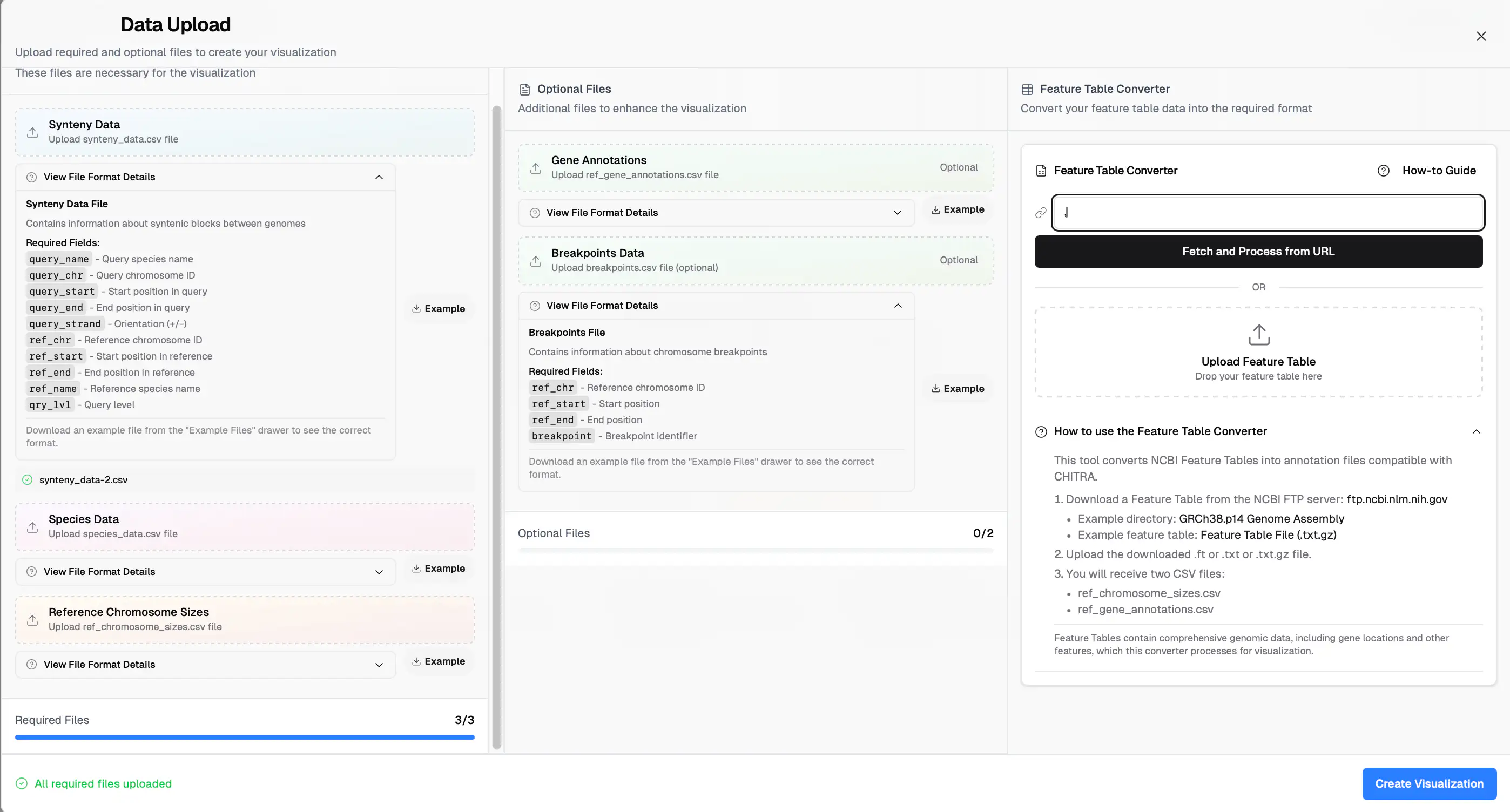
2.1.1 File Upload Instructions
Mandatory Input Files:
- Synteny Data
- Species Data
- Reference Chromosome Size
Optional Input Files:
- Gene Annotations
- Breakpoint Data
To upload files:
Click the “Upload Data” button. A popup window will appear where you can browse and upload the required input files.
Note: All input files should be in CSV format
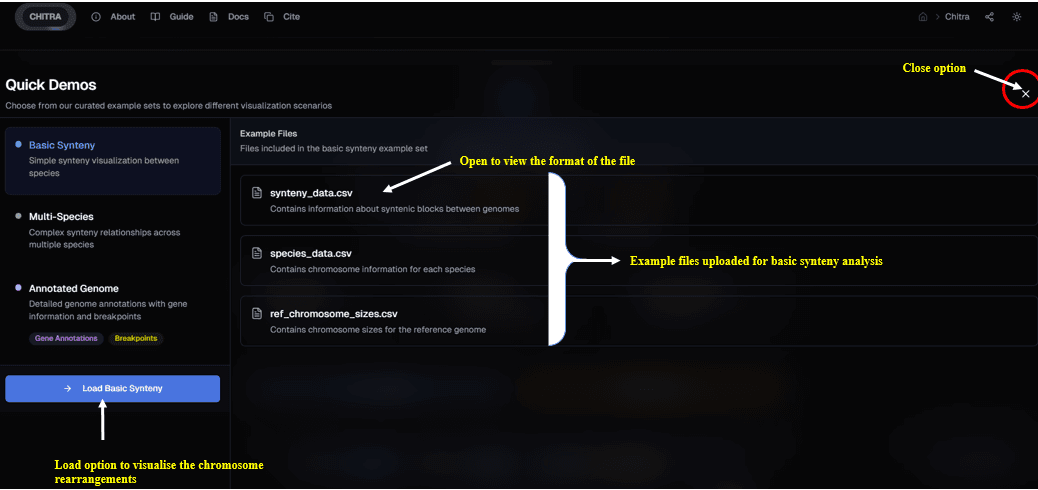
2.1.2 Using Example Datasets
CHITRA includes pre-loaded example datasets to help users explore its functionality. Three types of example datasets are available:
1. Basic Synteny
Visualize synteny blocks among three genomes.
Input files required:
2. Multiple Synteny
Compare synteny blocks among multiple genomes. Uses the same files as Basic Synteny.
3. Annotated Genome
Analyze annotated genomes using real datasets. Uses the same files as Basic Synteny, plus these additional files:
Available Datasets
Three types of example datasets are available:
1. Basic Synteny
- Simple synteny visualization between species.
- Ideal for understanding the fundamental concepts of synteny blocks and chromosome mapping.
2. Multi-Species
- Complex synteny relationships across multiple species.
- Allows comparison of genomic architecture across diverse organisms.
- Includes data for multiple pairwise comparisons.
3. Annotated Genome
- Detailed genome annotations with gene information and breakpoints.
- Includes gene features, locus tags, and symbols.
- Provides breakpoint data for structural variation analysis.
[!NOTE] You can preview and download the example data tables directly from the example files drawer below.
Exploring Example Data
- Select any of the three options in the drawer to access the files.
- You can preview the data or download the files directly.
- Click "Load" to automatically populate the visualization with the selected dataset.
Feature Table Converter
This tool converts NCBI Feature Tables into annotation files compatible with CHITRA.
How to use the Feature Table Converter
-
Download a Feature Table
- Get the file from the NCBI FTP server.
- Example directory: GRCh38.p14 Genome Assembly
- Example file: Feature Table File (.txt.gz)
-
Upload the File
- Upload the downloaded
.ft,.txt, or.txt.gzfile to the converter.
- Upload the downloaded
-
Download Converted Files
- You will receive two CSV files compatible with CHITRA:
ref_chromosome_sizes.csv: Contains the size of each chromosome.ref_gene_annotations.csv: Contains detailed gene annotation data.
- You will receive two CSV files compatible with CHITRA:
Note: Feature Tables contain comprehensive genomic data, including gene locations and other features, which this converter processes for visualization.